Research
Our research, mission and vision
Background information
The department Molecular Genetics is headed by Roland Kanaar and focuses on how the integrity of DNA is maintained by the DNA damage response, a process central to the present-day problems of cancer and aging-related diseases.

Our department has a long history and world-wide reputation in understanding mechanisms of the DNA damage response from defined molecular level to the complex context of physiological processes. Its fundamental studies of molecular mechanisms provide the basis for the rational design and development of novel targeted therapies and evidence-based sustainment of health.
Our ambitions
- Address the fundamental molecular mechanistic aspects of the DNA damage response by pursuing complementary and interwoven approaches at molecular and cellular levels.
- Provide a rational basis for improving existing approaches and developing novel and innovative precision therapies for cancer and aging-related diseases.
- Bring to the clinic better criteria and tools for individual patient selection for precision therapy concerning cancer.
- Develop reliable prognostic tools for treatment of cancer and aging-related diseases. Educate and train the next generation of multidisciplinary life scientists and healthcare professionals.

Our mission
We generate new and fundamental mechanistic knowledge on the DNA damage response through multi-disciplinary research. As we collaborate with our colleagues in the clinic, we aim to design novel targeted therapies to increase the quality of life of cancer patients and to increase the healthy lifespan of an aging population.
Principal Investigators
 |
DNA Repair Research for Optimized Tumor Treatment |
 |
Aging |
 |
Chronobiology and Health |
 |
Homologous Recombination and DSB metabolism |
 |
DNA Mismatch Repair |
 |
Transcription Stress and Genome Stability |
 |
Radiobiology of targeted radionuclide therapy |
 |
Single Cell Technology for Cancer Biology |
 |
DNA replication stress and genome instability |
 |
Nucleotide Excision Repair |
 |
Chromatin Remodeling and Replication |
 |
DNA repair mechanisms and disease |
 |
Alex Pines
Transcription interfering lesions and cellular homeostasis |
 |
Optogenetics and Imaging Methods |
 |
Molecular Mechanisms of Cardiovascular Aging |
 |
In vivo molecular imaging of Cancer and Cardiovascular disease |
 |
Molecular pathogenesis of neurodegenerative diseases |
Research Lines
The central research focus of the Department of Molecular Genetics at the Erasmus University Medical Center Rotterdam is the DNA damage response. We analyze this using an integrated approach from the molecular level to the physiological processes in the body, in the context of cancer, aging and aging-related diseases.
The four research lines underlying the central theme are:
- Molecular Mechanisms of the DNA Damage Response
- Cellular Function of the DNA Damage Response
- The DNA Damage Response in Cancer
- The DNA Damage Response in Aging
The Department of Molecular houses 18 Principal Investigators contributing to the four research lines. The listed distribution of Principal Investigators over the research lines is fluid, due to intradepartmental collaborations and the fact that projects often span different research lines.
Research Line 1 Molecular Mechanisms of the DNA Damage Response
This research line addresses the fundamental molecular mechanistic aspects of the DNA damage response, with a focus on homologous recombination, interstrand cross-link repair and mismatch repair. The molecular mechanisms are studied by applying and developing (single molecule) biochemical and biophysical techniques and analyses, advanced proteomic analyses and state-of-the-art molecular imaging techniques to identify the dynamic molecular interactions that are responsible for the assembly and disassembly of the molecular factories that guard and repair the genome.
More specifically, we:
- Quantitatively describe BRCA2 protein dynamics (time, space, shape changes) using an ensemble of techniques including scanning force microscopy (SFM), total internal reflection fluorescence (TIRF)-SFM, and force spectroscopy.
- Understand the mode of action and function of internally-disordered regions in BRCA2 in the context of its interaction with RAD51.
- Characterize and understand the function of novel components identified in the BRCA2 interactome.
- Quantitatively describe the mismatch DNA repair pathway (recognition, communication along DNA, excision, repair synthesis) through the integration of results from biochemical and biophysical experiments and computer simulation.
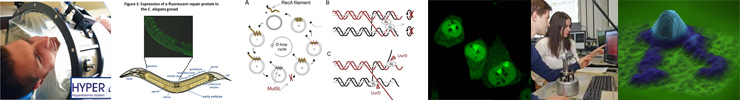
The ultimate goal is to define and dissect the molecular circuits of DNA damage repair and their integration in cellular processes (see Research Line 2, below). This knowledge is pivotal for identifying molecular targets for designing novel mechanism-based interventions and manipulations of these processes. This will provide the basis for improving existing approaches and developing entirely novel and innovative precision therapies for cancer and aging-related diseases.
Research Line 2 Cellular Function of the DNA Damage Response
At the cellular level, the DNA damage repair processes in (live) cells are monitored in real time to see how they detect DNA damage and proceed to assemble and disassemble the repair machinery. We apply the latest super-resolution techniques and single particle tracking analyses to understand the interplay between different DNA damage response pathways and their integration in the physiology of the cell.
More specifically, we:
- Identify the causes and functional consequences of transcription inhibition due to DNA damage and of cellular responses to transcription stress.
- Dissect the functional interplay between DNA repair and chromatin remodeling in genome stability, cancer, chemo-resistance, and (therapy-induced) DNA replication stress.
- Understand the mechanisms underlying replication fork protection and its role in the prevention of tumorigenesis.
- Systemically and functionally characterize DNA damage responses towards chemotherapy with the (ultimate) aim to improve cancer treatment.
- Understand tissue-specific DNA repair organization using in vitro and in vivo differentiation models (iPS cells) to move towards a full comprehension of the pathogenesis of DNA repair disorders.
- Establish high quality standards of DNA repair assays and genotyping for the research-based diagnostics of DNA repair disorders.
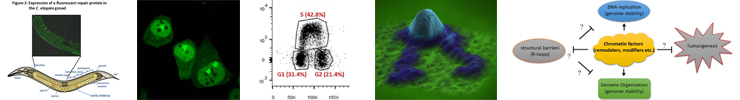
- Jurgen Marteijn
- Pier Giorgio Mastroberardino
- Hannes Lans
- Arnab Ray Chaudhuri
- Nitika Taneja
- Maria Tresini
- Wim Vermeulen
Research Line 3 The DNA Damage Response in Cancer
Many important anti-cancer treatments are based on the cell-killing properties of DNA-damaging agents. The efficacy of such treatments depends on the characteristics of the DNA damage response in tumour cells. The DNA damage response is almost invariably compromised in tumours, and this can be exploited to develop better patient selection methods, guide targeted cancer treatments, and design novel precision cancer treatments. Based on extensive analysis of the DNA damage response in cell culture, the department has developed a clinic-to-lab pipeline for breast cancer patients. This allows testing, in a functional ex vivo assay, aspects of the DNA damage response in viable (organotypic) slices of tumour material from individual patients.
More specifically, we:
- Identify cellular pathways involved in the repair of DNA damage induced by different radiation qualities (α-radiation and β-radiation versus X-rays) and the biological effectiveness of the radiation.
- Increase molecular targeted radionuclide therapy efficacy through a combination with DNA repair inhibitors.
- Develop functional assays to predict the therapy response to personalized medicine in cancer patients.
- Develop Cancer-on-Chip approaches to study therapy responses in real time.
- Develop next-generation, high-throughput screening microscopy technology for single cell applications to investigate the genetic, proteomic and transcriptomic features of specially-selected (stem-like) cancer cells in a population.
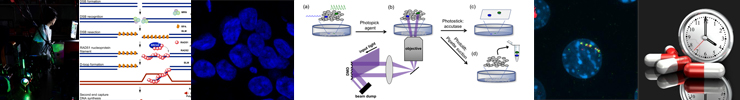
Research Line 4 The DNA Damage Response in Aging
DNA damage does not just foster mutagenesis and thereby carcinogenesis, it also constitutes a major cause of cellular dysfunction, cell death and cellular senescence – all of which drive the aging process. As a result, many inborn disorders due to genetic deficiencies in DNA damage response pathways exhibit a strong cancer predisposition and/or premature aging symptoms. The department has invested in generating a comprehensive series of mouse mutants carrying engineered defects in specific repair genes, some of which precisely mimic the defect of DNA repair-deficient patients. These mouse mutants turn out to be extremely informative – they mimic the corresponding human syndromes and also enable detailed insight into the complex etiology of human repair syndromes. This strong connection between accumulating DNA damage and accelerated – but truly bona fide aging – was discovered as a trade-off between cancer and aging. Our collection of genetically-engineered progeroid mouse mutants, which is unique in the world, provides the department with an exclusive competitive advantage. We are now using them to develop intervention strategies for the diseases associated with aging-related symptoms.
More specifically, we:
- Elucidate the role of DNA damage and the associated transcription stress in the aging process.
- Identify how nutrition and anti-aging interventions control DNA damage load, DNA damage repair and signalling.
- Identify how DNA damage leads to age-related diseases such as dementia, Parkinson’s disease and cardiovascular diseases.
- Elucidate how DNA damage impinges on disease mechanisms such as loss of proteostasis, senescence and stem cells.
- Develop genomics tools and software to understand DNA damage accumulation and DNA repair in aging at nucleotide resolution and the single cell level.
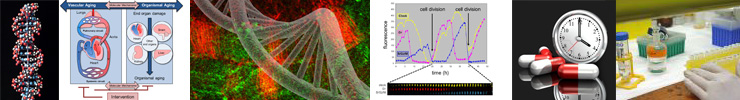
More information can be found at:
Projects
Notable results
Breakthrough discoveries
- First molecular cloning of a chromosome translocation break point in cancer, providing insight in the molecular basis of cancer and enabling development of the first cancer type specific drug (Imatanib/Gleevec)
- Molecular cloning of the first mammalian DNA repair gene and discovery of the very strong evolutionary conservation of DNA repair
- Discovery of the connection between transcription and DNA repair and identification of human transcription syndromes
- Discovery of the role of mammalian photolyase paralogs in the circadian/biological clock
- Discovery of the connection between the DNA damage response
- Identification of nutritional interventions for the treatment of human DNA repair syndromes exhibiting accelerated aging
- Discovery of the pathways of random DNA integration in mammalian genomes
Publications
Please visit Pure for all our Research output
Our key papers:
M. Tresini, D.O. Warmerdam, P. Kolovos, L. Snijder, M.G. Vrouwe, J.A. Demmers, W.F. van IJcken, F.G. Grosveld, R.H. Medema, J.H. Hoeijmakers, L.H. Mullenders, W. Vermeulen and J.A. Marteijn.
The core spliceosome as target and effector of non-canonical ATM signaling.
Nature, 523: 53-58 (2015)
doi: 10.1038/nature14512. PMID: 26106861
C. Dinant, G. Ampatziadis-Michailidis, H. Lans, M. Tresini, A.F. Theil, W.A. van Cappellen, H. Kimura, J. Bartek, M. Fousteri, A.B. Houtsmuller, W. Vermeulen and Marteijn, J.A.
Enhanced chromatin dynamics by FACT promotes transcriptional restart after DNA damage.
Molecular Cell 51: 469-79 (2013).
doi: 10.1016/j.molcel.2013.08.007. PMID: 23973375
P. Schwertman, A. Lagarou, D.H. Dekkers, J. Raams, A.C. van der Hoek, C. Laffeber, J.H. Hoeijmakers, J.A. Demmers, M. Fousteri, W. Vermeulen and J.A. Marteijn.
UV-sensitive syndrome protein KIAA1530 recruits USP7 to regulate transcription coupled repair.
Nature Genetics 44: 598-602 (2012).
doi: 10.1038/ng.2230. PMID: 22466611
P.M. Krawczyk, B. Eppink, J. Essers, J. Stap, H. Rodermond, H. Odijk, A. Zelensky, C. van Bree, L.J. Stalpers, M.R. Buist, T. Soullié, J. Rens, H.J. Verhagen, M.J. O'Connor, N.A. Franken, T.L. ten Hagen, R. Kanaar and J.A. Aten.
Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition.
Proc. Natl. Acad. Sci. USA 108, 9851-9856 (2011).
doi: 10.1073/pnas.1101053108. PMID: 21555554
M. Modesti, M. Budzowska, C. Baldeyron, J.A. Demmers, R. Ghirlando and R. Kanaar. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination.
Molecular Cell 28, 468-481 (2007).
PMID: 17996710
K. Hanada, M. Budzowska, S.L. Davies, E. van Drunen, B.H. Beverloo, A. Maas, J. Essers, I.D. Hickson and R. Kanaar.
The structure-specific endonuclease Mus81-Eme1 contributes to replication restart by generating double-strand DNA breaks.
Nat. Struct. Mol. Biol. 14, 1096-1104 (2007).
PMID: 17934473
L.J. Niedernhofer, G.A. Garinis, A. Raams, S.A. Lalai, R.A. Robinson, E. Appeldoorn, H. Odijk, R. Oostendorp, A. Ahmad, W. van Leeuwen, A. Theil, W. Vermeulen, G.T. van der Horst, P. Meinecke, W. Kleijer, J. Vijg, N.G.J. Jaspers, J.H.J. Hoeijmakers.
A new progeriod syndrome reveals that genotoxic stress suppresses the somatotroph axis.
Nature 444: 1038-1043 (2006).
PMID: 17183314
Giglia-Mari, G., Coin, F., Ranish, J.A., Hoogstraten, D., Theil, A., Wijgers, N., Jaspers, N.G., Raams, A., Argentini, M., van der Spek, P.J., Botta, E., Stefanini, M., Egly, J.M., Aebersold, R., Hoeijmakers, J.H., and Vermeulen, W.
A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A.
Nature Genetics 36: 714-719 (2004).
PMID: 15220921
J.A. Aten, J. Stap, P.M. Krawczyk, C.H. van Oven, R.A. Hoebe, J. Essers and R. Kanaar.
Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains.
Science 303, 92-95 (2004).
PMID: 14704429
Hoogstraten, D., Nigg, A.L., Heath, H., Mullenders, L.H., van Driel, R., Hoeijmakers, J.H., Vermeulen, W. and Houtsmuller, A.B.
Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo.
Molecular Cell 10: 1163-1174 (2002)
PMID: 12453423
J. de Boer, J.O. Andressoo, J. de Wit, J. Huijmans, R.B. Beems, H. van Steeg, G. Weeda, G.T.J. van der Horst, W. van Leeuwen, A.P.N. Themmen, M. Meradji and J.H.J. Hoeijmakers.
Premature aging in mice deficient in DNA repair and transcription.
Science (research article), 296: 1276-1279 (2002).
PMID: 11950998
W. Vermeulen, S. Rademakers, N.G.J. Jaspers, E. Appeldoorn, A. Raams, B. Klein, W. Kleijer, L. Kjærsgård and J.H.J. Hoeijmakers.
A temperature-sensitive disorder in basal transcription and DNA repair in man.
Nature Genetics 27: 299-303 (2001).
PMID: 11242112
M. de Jager, J. van Noort, D.C. van Gent, C. Dekker, R. Kanaar and C. Wyman.
Human Rad50/Mre11 is a flexible complex that can tether DNA ends.
Molecular Cell 8: 1129-1135 (2001).
PMID: 11741547
Vermeulen, W., Bergmann, E., Auriol, J., Rademakers, S., Frit, P., Appeldoorn, E., Hoeijmakers, J.H., and Egly, J.M.
Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder.
Nature Genetics 26: 307-313 (2000).
PMID: 11062469
A.B. Houtsmuller, S. Rademakers, A.L. Nigg, D. Hoogstraten, J.H.J. Hoeijmakers, W. Vermeulen.
Action of DNA repair endonuclease ERCC1/XPF in living cells.
Science 284: 958-961 (1999).
PMID: 10320375
H. Okamura, S. Miyake, Y. Sumi, S. Yamaguchi, A. Yasui, M. Muijtjens, J.H.J. Hoeijmakers and G.T.J. van der Horst.
Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock.
Science 286: 2531-2534 (1999).
PMID: 10617474
G.T.J. van der Horst, M. Muijtjens, K. Kobayashi, R. Takano, S-I. Kanno, M. Takao, J. de Wit, A. Verkerk, A.P.M. Eker, D. van Leenen, R. Buijs, D. Bootsma, J.H.J. Hoeijmakers, A. Yasui.
Mammalian blue-light receptor homologs CRY1 and CRY2 are essential for maintenance of the biological clock.
Nature 398: 627-630 (1999).
PMID: 10217146
K. Sugasawa, J.M.Y. Ng, C. Masutani, P.J. van der Spek, A.P.M. Eker, F. Hanaoka, D. Bootsma and J.H.J. Hoeijmakers.
Xeroderma pigmentosum group C complex is the initiator of global genome repair.
Molecular Cell 2: 223-232 (1998).
PMID: 9734359
G.T.J. van der Horst, H. van Steeg, R.J.W. Berg, A.J. van Gool, J. de Wit, G. Weeda, H. Morreau, R.B. Beems, C.F. van Kreijl, F.R. de Gruijl, D. Bootsma and J.H.J. Hoeijmakers.
Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition.
Cell 89: 425-435 (1997).
PMID: 9150142
J. Essers, R.W. Hendriks, S.M.A. Swagemakers, C. Troelstra, J. de Wit, D. Bootsma, J.H.J. Hoeijmakers and R. Kanaar.
Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination.
Cell 89: 195-204 (1997).
PMID: 9108475
H. Roest, J. van Klaveren, J. de Wit, C.G. van Gurp, M.H.M. Koken, M. Vermey, J.H. van Roijen, J.T.M. Vreeburg, W.M. Baarends, D. Bootsma, J.A. Grootegoed and J.H.J. Hoeijmakers.
Inactivation of a ubiquitin-conjugating DNA repair enzyme in mice causes a defect in spermatogenesis associated with chromatin modification.
Cell 86: 799-810 (1996).
PMID: 8797826
Schaeffer, L., Roy, R., Humbert, S., Moncollin, V., Vermeulen, W., Hoeijmakers, J.H., Chambon, P. and Egly, J.M. (1993).
DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor.
Science 260: 58-63 (1993).
PMID: 8465201
Troelstra, A. van Gool, J. de Wit, W. Vermeulen, D. Bootsma and J.H.J. Hoeijmakers.
ERCC6, a member of a subfamily of putative helicases is involved in Cockayne's syndrome and preferential repair of active genes.
Cell 71: 939-953 (1992).
PMID: 1339317
G. Weeda, R.C.A. van Ham, W. Vermeulen, D. Bootsma, A.J. van der Eb and J.H.J. Hoeijmakers.
A presumed DNA helicase, encoded by the excision repair gene ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome.
Cell 62: 777-791 (1990).
PMID: 2167179
M. van Duin, J. de Wit, H. Odijk, A. Westerveld, A. Yasui, M. Koken, J.H.J. Hoeijmakers and D. Bootsma.
Molecular characterization of the human excision repair gene ERCC-1: cDNA cloning and aminoacid homology with the yeast DNA repair gene RAD10.
Cell 44: 913-923 (1986).
PMID: 2420469
Westerveld, J.H.J. Hoeijmakers, M. van Duin, J. de Wit, H. Odijk, A. Pastink, R.D. Wood and D. Bootsma.
Molecular cloning of a human repair gene.
Nature 310: 425-429 (1985).
PMID: 6462228

Shared facilities
Research facilities
The department makes use of the following Erasmus MC core facilities:
- Applied Molecular Imaging Erasmus MC (AMIE): AMIE offers multimodal and high resolution in vivo imaging of small animals and tissue. We analyze genetically-engineered mouse models in cancer and aging research.
- Erasmus Center for Animal Research: We generate and house mouse strains.
- Erasmus Optical Imaging Center: An expert center for advanced optical imaging knowledge and equipment. We conduct cell biological analyses of the DNA damage response.
- ERGO: A prospective cohort study on the risk factors and determinants of chronic diseases in middle-aged and elderly persons. We identify circadian disturbance-related epigenetic risk markers for aging related pathologies.
- Generation R: A population-based prospective cohort study from fetal life until adulthood. We identify pre- and early postnatal circadian disturbance-related epigenetic risk markers for later life disease.
- Genomics: Expertise Center for Genomics Research. We conduct sequence-based experiments on the effect of the DNA damage response in cancer and aging at Erasmus Center for Biomics and and HuGe-F.
- iPS facility: We conduct the derivation and distribution of iPSCs for human disease modelling. We derive iPS cells from patients with a DNA damage response defect. We also use it as a user facility location and to conduct onsite training.
- Erasmus MC Proteomics Center: Mass spectrometry-based proteomics services for the Erasmus MC community and external researchers. We conduct biochemical analyses of protein complexes involved in the DNA damage response.





















































