About Dr. J. (Joyce) Lebbink, PhD
Introduction
About Joyce
Joyce Lebbink studied Molecular Sciences (specialty Biochemistry) at the Wageningen University and Research Center in the Netherlands from 1988 to 1994. She obtained her PhD in 1999 at the same University, characterizing molecular details of thermostable enzymes at the Department of Microbiology. She further obtained experience in biophysics and structural biology during her Postdoc at the Center for Structural Biochemistry, Karolinska Institute in Sweden. In 2002 she returned to the Netherlands as Postdoc in the Protein Crystallography group of Prof. Titia Sixma at the Netherlands Cancer Institute to study structure and mechanisms of DNA Mismatch Repair.

Since September 2007 she is further studying mismatch repair and its interactions with homologous recombination at the Department of Molecular Genetics at the Erasmus MC.
Joyce has received Marie-Curie, NWO-VENI and NWO-VIDI personal fellowships and is scientific coordinator of the EU-Marie Curie Training Network “DNARepairMan”.
Scientific Coordination
DNARepairMan

Joyce is Scientific Coordinator of the European Training Network DNARepairMan. DNARepairMan is an innovative training network aimed at understanding the regulated assembly of molecular machines for DNA repair. The Network aims to provide the next generation of young scientists with a unique and innovative interdisciplinary training in cutting-edge biophysical research methodologies to address central questions in biology with relevance for human health. The Network is composed of 7 academic laboratories, 4 companies and an Art Academy.
Mismatch2model

Joyce was Scientific Coordinator of the EU-FP7-HEALTH collaborative project ‘mismatch2model: Characterization and quantitative modeling of DNA mismatch repair and its role in the maintenance of genomic stability and cancer avoidance’, from November 2008 – October 2012. This multidisciplinary team of biochemists, biophysicists, structural biologists, cell biologists and quantitative modelers is studied the molecular mechanisms of DNA mismatch repair with a multidisciplinary approach.
Field(s) of expertise
Molecular Mechanisms of DNA Mismatch Repair
DNA mismatch repair (MMR) is responsible for correcting DNA replication errors and controls illegitimate DNA recombination. Non-functional MMR results in a mutator phenotype and in humans in an increased risk to develop cancer. MMR is a highly coordinated, multistep pathway in which a correct communication between the involved proteins on DNA is of crucial importance. Using in vitro reconstituted MMR reactions, we study this regulation in molecular detail.

Research interests
DNA mismatch repair (MMR) corrects DNA replication errors and inhibits recombination between DNA with diverging sequences. In both bacteria and eukaryotes, the loss of mismatch repair gives rise to a mutator phenotype. In the most frequent form of familiar cancer, Lynch syndrome, germline mutations in one of the DNA mismatch repair genes predispose to cancer of the colon, endometrium, ovary and other organs.
We would like to understand in mechanistic detail how the recognition of a DNA mismatch during DNA replication or in recombination intermediates results in intramolecular signaling, repair complex formation and ultimately, correct mismatch repair or abortion of the recombination event.
We approach these research questions using a combination of molecular biology, quantitative biochemistry, biophysics and structural biology. The importance of these kind of studies was emphasized by the award of the 2015 Chemistry Nobel prize to three pioneers of the field for their mechanistic studies of DNA Repair. We have long-standing collaborations on this subject within the Netherlands (Titia Sixma at NKI) and abroad (Peter Friedhoff in Giessen, Germany; Terence Strick at CNRS, Paris).
Expertise
- Protein expression and purification
- Surface plasmon resonance (kinetics of protein-protein and protein-nucleic acids interactions)
- Functional biochemical assays (enzyme activity, DNA binding, DNA incision, DNA unwinding, DNA excision)
- Fluorescence spectroscopy (anisotropy and FRET)
- Protein structure analysis

Current members
Charlie Laffeber, technical expert
Yannicka Mardenborough, PhD student
Enes Sahin, M.Sc student Nanobiology
(pictures)
Alumni
Pauline Ikpa, technician
Michele Cristovao, postdoc
Nicolaas (Klaas) Hermans, PhD
Khek-Chian (KC) Tham, PhD
Publications
Selected Publications
Brandsma I, Sato K, van Rossum-Fikkert SE, van Vliet N, Sleddens E, Reuter M, Odijk H, van den Tempel N, Dekkers DHW, Bezstarosti K, Demmers JAA, Maas A, Lebbink J, Wyman C, Essers J, van Gent DC, Baarends WM, Knipscheer P, Kanaar R, Zelensky AN. HSF2BP Interacts with a Conserved Domain of BRCA2 and Is Required for Mouse Spermatogenesis. Cell Rep. 27: 3790-3798. 2019.
Mardenborough YSN, Nitsenko K, Laffeber C, Duboc C, Sahin E, Quessada-Vial A, Winterwerp HHK, Sixma TK, Kanaar R, Friedhoff P, Strick TR, Lebbink JHG. The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair. bioRxiv 463133; doi: https://doi.org/10.1101/463133.
Wu WY, Lebbink JHG, Kanaar R, Geijsen N, van der Oost J. Genome editing by natural and engineered CRISPR-associated nucleases. Nature Chem. Biol. 14, 642-651, 2018.
Sanchez H, Paul MW, Grosbart M, van Rossum-Fikkert SE, Lebbink JHG, Kanaar R, Houtsmuller AB and Claire Wyman C. Architectural plasticity of human BRCA2–RAD51 complexes in DNA break repair. Nucleic Acids Res. 45, 4507-4518, 2017.
Hermans N, Laffeber C, Cristovão M, Artola-Borán M, Mardenborough Y, Ikpa P, Jaddoe A, Winterwerp HHK, Wyman C, Jiricny J, Kanaar R, Friedhoff P, Lebbink JHG. Dual daughter strand incision is processive and increases the efficiency of DNA mismatch repair. Nucleic Acids Res. 44, 6770-6778, 2016.
Yu N, Signorile L, Basu S, Ottema S, Lebbink JH, Leslie K, Smal I, Dekkers D, Demmers J, Galjart N. Isolation of functional tubulin dimers and of tubulin-associated proteins from mammalian cells. Curr Biol. 2016 S0960-9822(16)30463-8. doi: 10.1016/j.cub.2016.04.069, 2016.
Tham KC, Kanaar R, Lebbink JHG. Mismatch repair and homeologous recombination. DNA Repair 38, 75-83, 2016.
Groothuizen FS, Winkler I, Cristóvão M, Fish A, Winterwerp HHK, Reumer A, Marx AD, Hermans N, Nicholls RA, Murshudov GN, Lebbink JHG, Friedhoff P, Sixma TK. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife 10.7554/eLife.06744, 2015.
Tham KC, Hermans N, Winterwerp HHK, Cox MC, Wyman C, Kanaar R and Lebbink JHG. Mismatch Repair Inhibits Homeologous Recombination via Coordinated Directional Unwinding of Trapped DNA Structures. Mol. Cell. 51, 326-337, 2013.
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HHK, Marinus MG, Lebbink JHG, Svergun DI, Friedhoff P and Sixma TK. Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation. Nucleic Acids Res. 14, 8166-8181, 2013.
Teaching activities
B.Sc. Nanobiology:
Elective “Protein Structure: Theory and Tools”
M.Sc. Nanobiology:
Engineering Genetic Information
Ph.D. Biomedical Sciences:
Special Topics Biochemistry and Biophysics

More
Research
In Escherichia coli, MMR is initiated by MutS and MutL, which in an ATP-dependent manner activate MutH to incise transiently-hemimethylated GATC sites on the nascent DNA strand containing the replication error. MutL then recruits the helicase UvrD which unwinds the DNA from the nick towards the mismatch, after which exonucleases degrade the displaced strand and the replication machinery restores the missing sequence.
Projects
Processivity during DNA MMR
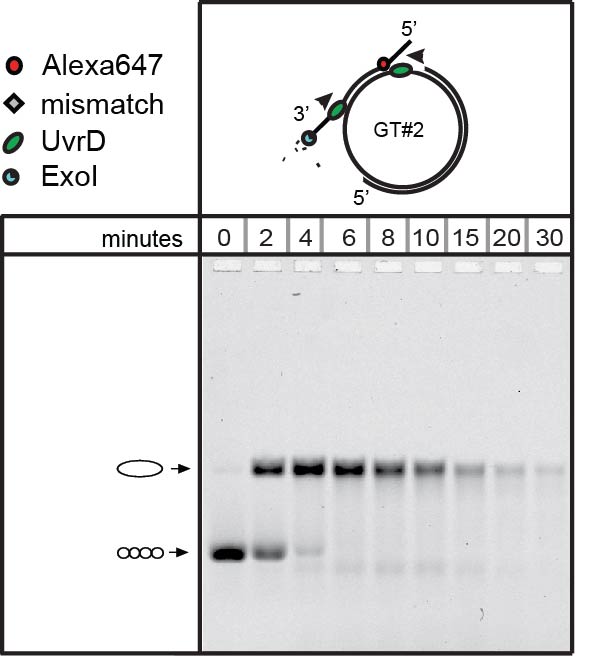
MMR efficiency depends on the distribution of the hemimethylated GATC sites. We found that two GATC sites were incised by the same activated MMR complex in a processive manner, with MutS, the closed form of MutL and MutH displaying different roles. Unwinding and strand excision were more efficient on a substrate with two nicks flanking the mismatch, as compared to substrates containing a single nick or two nicks on the same side of the mismatch (Hermans et al., 2016). We are currently further investigating the details of the different roles of the MMR proteins in this process.
Timing of events
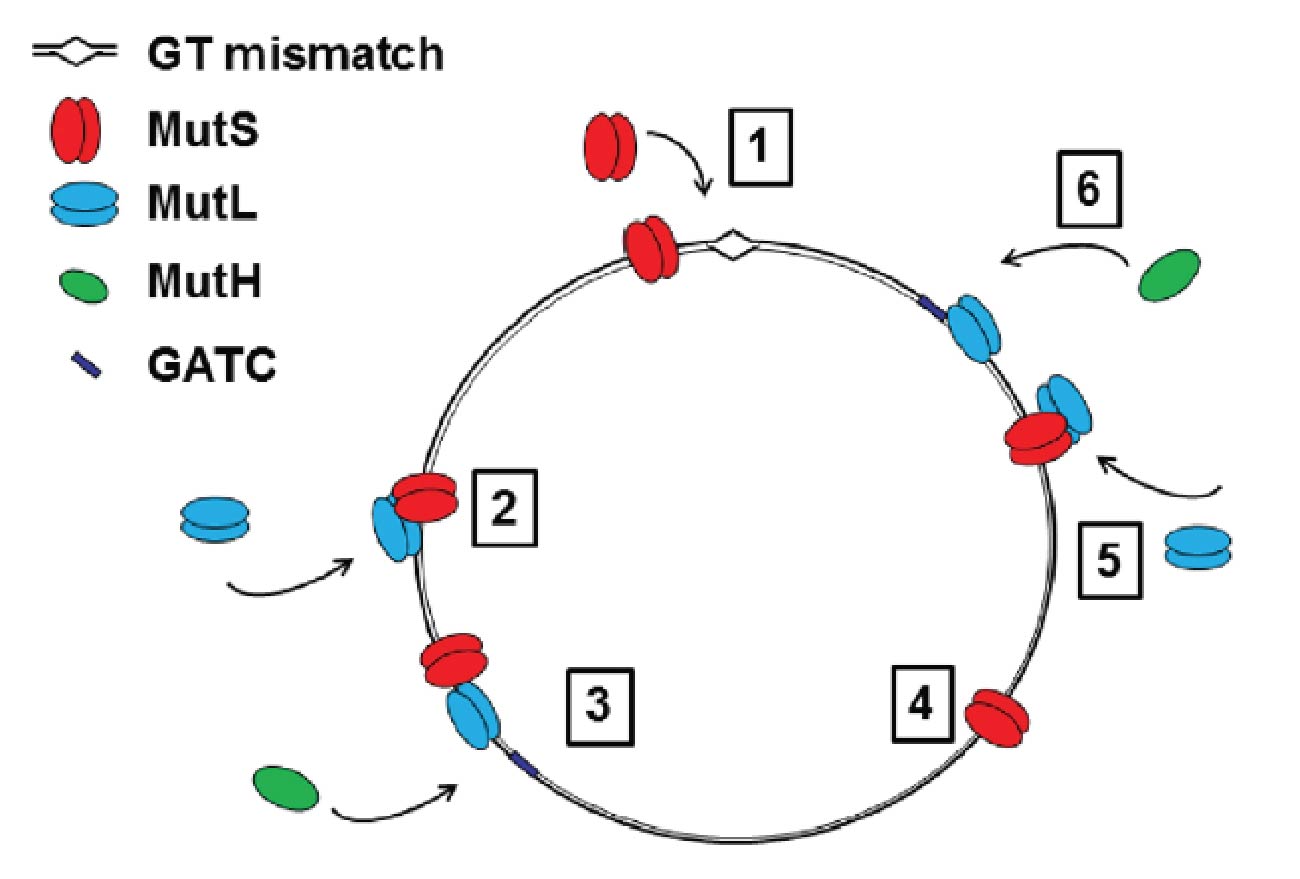
Processive strand incision by MutH requires activated MutS and the activated, closed state of MutL. By varying the order of addition of MutS, MutL, MutH and ATP to allow pre-loading of activated MMR complexes, and combined with Monte Carlo simulations, we deduced that MutL activation is rate-limiting for incision. In collaboration with the laboratory of Peter Friedhoff in Giessen we are currently verifying this finding using fluorescence-based conformational assays. We will furthermore analyse the effects of specific mutations in MutS and MutL on protein conformation and activity. These experiments will correlate the extent of activation of the MMR complex in different steps of the reaction to the predominant conformational states of MutS and MutL.
Mode of communication
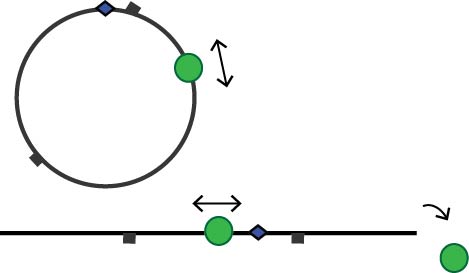
The mode of communication between mismatch and GATC site is an unresolved issue within the MMR field, with different models available in the literature, amongst which loop formation in the DNA and a diffusive search along the DNA backbone. A confounding factor in this controversy is the absence of correlative qualitative behaviour of protein-DNA complexes and quantitative functional analysis. In this sub-project we will correlate the processive nicking activity of the activated MMR complex with its dependence on the presence of DNA ends, MutS oligomerization and loop formation. We will combine this with Monte Carlo simulations to obtain more insight in this elusive process.
Directionality of MMR

Unwinding by the UvrD helicase during MMR proceeds from the strand incision created by MutH in the direction of the mismatch. As the helicase can be loaded at the nick onto either DNA strand, the MutSL complex or a specific signal on the DNA must provide the information that controls directional loading of the helicase. In this project we will unravel the molecular details of this directionality.
Antirecombination
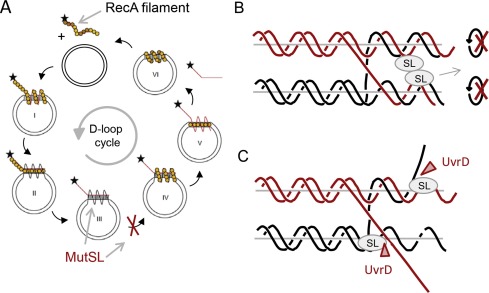
Homeologous recombination between divergent DNA sequences is inhibited by DNA mismatch repair. In Escherichia coli, MutS and MutL respond to DNA mismatches within recombination intermediates and prevent strand exchange. We recently established that in addition to mismatches within the heteroduplex region, secondary structures within the displaced ssDNA formed during branch migration within the recombination intermediate are involved in the inhibition. We present a model that explains how higher-order complex formation of MutS, MutL and DNA blocks branch migration by preventing rotation of the DNA strands within the recombination intermediate. Furthermore, we found that the helicase UvrD is recruited to directionally resolve these trapped intermediates toward DNA substrates (Tham et al, 2013). In this project we will extend this analysis to the eukaryotic systems.
Student projects
We have M.Sc. End Projects available. Check the research descriptions above.


