About Dr. K.S. (Kerstin) Wendt
Field(s) of expertise
Cohesin and chromatin organization
The activity of genes depends on regulatory elements such as promoters and enhancers, but also on the folding of the chromatin fibre, which in one case can promote the interactions between genes and their regulatory elements and in other cases blocks these interactions, leading to gene silencing.
Our research interest is to understand which proteins are involved in shaping the 3D-architecture, in particular of the human genome. We are in particular interested in the cohesin complex and the chromatin insulator protein CTCF.
Cohesin is an important factor to tether DNA strands during cell divisions, for DNA damage repair and as we could show to promote long-range interactions between distant regions of the DNA-strand, also referred to as chromatin loops. Genome-wide cohesin largely colocalizes and cooperates with the chromatin insulator CTCF and to some extend also with other transcription factors like the mediator complex.
We are using different methods to visualize the interactions of the chromatin fibre in human cell lines but also primary tissues: different chromatin conformation capturing methods like 3C-seq (similar to 4C); Hi-C and the novel high-resolution method Targeted Chromatin Conformation Capturing (T2C) that we have developed together with other groups at the Erasmus MC. To determine, which proteins bind and where we use chromatin immunoprecipitation (ChIP) in combination with mass spectrometry (ChIP-MS) and next generation sequencing (ChIP-seq).
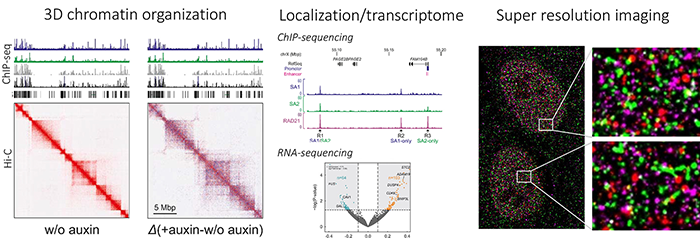
Cohesin-related diseases and developmental syndromes
Our second research interest is how mutations linked to cohesin can lead to development and defects and also cancer. Mutations of the cohesin complex, but also a number of its regulators are found in several human developmental syndromes like Cornelia de Lange Syndrome and Roberts SC phocomelia syndrome. To investigate these syndromes we are tightly collaborating with research groups in Germany and in France in the collaborative network Target-CdLS.
Education and career
During her PhD as structural biologist Kerstin S. Wendt solved the first protein crystal structure of a component of the APC/C complex, an important cell cycle regulator. This work raised her interest in cell cycle regulators and in particular in proteins that can organize DNA strands. During her PostDoc in the group of Jan-Michael Peters at the IMP in Vienna/Austria she discovered a novel condensin complex (condensin II). Further she determined in one of the first genome-wide mapping studies the position of the cohesin complex in the human genome. These studies lead to the discovery that the cohesin complex, well known to promote chromosome cohesion, cooperates with the chromatin insulator CTCF for chromatin insulation and transcriptional regulation. Shortly after she could show that the cohesin complex is involved in promoting long-range interactions of the chromatin fibre.
With her own research group, which she started in 2009 at the Erasmus MC, she investigates, which roles cohesin and CTCF have for shaping the overall structure of the chromatin fibre. Among the approaches used in her group are different chromatin conformation capturing techniques and imaging techniques such as DNA-FISH. The group is together with other groups at the department of Cell biology involved in the development of novel high resolution capture techniques.
Next to the interest in chromatin organization Kerstin S. Wendt has a strong interested in the developmental syndrome Cornelia de Lange Syndrome which is associated with mutations in cohesin complex subunits or cohesin regulators. In a consortium with several international partners she investigates how these mutations lead to the malformations observed in this congenital disorder.
Contact: Kerstin S. Wendt
Telephone: +31-107044007
E-mail: k.wendt@erasmusmc.nl
Publications
Key publications:
Zuin J, Casa V, Pozojevic J, Kolovos P, van den Hout MCGN, van Ijcken WFJ, Parenti I, Braunholz D, Baron Y, Watrin E, Kaiser FJ, Wendt KS. Regulation of the cohesin-loading factor NIPBL: Role of the lncRNA NIPBL-AS1 and identification of a distal enhancer element. PLoS Genet. 2017 Dec 20;13(12):e1007137. doi: 10.1371/journal.pgen.1007137.
Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, Wendt KS, Okada Y, Nagai T, Maeshima K. Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging. Mol Cell. 2017 Jul 20;67(2):282-293.e7. doi: 10.1016/j.molcel.2017.06.018. Epub 2017 Jul 14.
Wendt KS. Resolving the Genomic Localization of the Kollerin Cohesin-Loader Complex. Methods Mol Biol. 2017;1515:115-123.
Wendt KS, Grosveld FG. Transcription in the context of the 3D nucleus. Curr Opin Genet Dev. 2014 Apr;25:62-7.
Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, Grosveld FG, Ren B, Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014 Jan 21;111(3):996-1001.
Zuin J, Franke V, van Ijcken WF, van der Sloot A, Krantz ID, van der Reijden MI, Nakato R, Lenhard B, Wendt KS. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 2014 Feb 13;10(2):e1004153.
Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009 Nov;5(11):e1000739.
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008 Feb 14;451(7180):796-801.
Full publication list:
2018
Kolovos P, Brouwer RWW, Kockx CEM, Lesnussa M, Kepper N, Zuin J, Imam AMA, van de Werken HJG, Wendt KS, Knoch TA, van IJcken WFJ, Grosveld F.
Investigation of the spatial structure and interactions of the genome at sub-kilobase-pair resolution using T2C.
Nat Protoc. 2018 Mar;13(3):459-477. doi: 10.1038/nprot.2017.132. Epub 2018 Feb 8.
2017
Zuin J, Casa V, Pozojevic J, Kolovos P, van den Hout MCGN, van Ijcken WFJ, Parenti I, Braunholz D, Baron Y, Watrin E, Kaiser FJ, Wendt KS.
Regulation of the cohesin-loading factor NIPBL: Role of the lncRNA NIPBL-AS1 and identification of a distal enhancer element.
PLoS Genet. 2017 Dec 20;13(12):e1007137. doi: 10.1371/journal.pgen.1007137.
Pozojevic J, Parenti I, Graul-Neumann L, Ruiz Gil S, Watrin E, Wendt KS, Werner R, Strom TM, Gillessen-Kaesbach G, Kaiser FJ.
Novel mosaic variants in two patients with Cornelia de Lange syndrome.
Eur J Med Genet. 2017 Nov 15. pii: S1769-7212(17)30498-6. doi: 10.1016/j.ejmg.2017.11.004.
Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, Wendt KS, Okada Y, Nagai T, Maeshima K.
Dynamic Organization of Chromatin Domains Revealed by Super-Resolution Live-Cell Imaging.
Mol Cell. 2017 Jul 20;67(2):282-293.e7. doi: 10.1016/j.molcel.2017.06.018. Epub 2017 Jul 14.
2016
Watrin E, Kaiser FJ, Wendt KS.
Gene regulation and chromatin organization: relevance of cohesin mutations to human disease.
Curr Opin Genet Dev. 2016 Apr;37:59-66. doi: 10.1016/j.gde.2015.12.004. Epub 2016 Jan 25.
Knoch TA, Wachsmuth M, Kepper N, Lesnussa M, Abuseiris A, Ali Imam AM, Kolovos P, Zuin J, Kockx CEM, Brouwer RWW, van de Werken HJG, van IJcken WFJ, Wendt KS, Grosveld FG.
The detailed 3D multi-loop aggregate/rosette chromatin architecture and functional dynamic organization of the human and mouse genomes.
Epigenetics Chromatin. 2016 Dec 24;9:58. doi: 10.1186/s13072-016-0089-x. eCollection 2016.
Parenti I, Gervasini C, Pozojevic J, Wendt KS, Watrin E, Azzollini J, Braunholz D, Buiting K, Cereda A, Engels H, Garavelli L, Glazar R, Graffmann B, Larizza L, Lüdecke HJ, Mariani M, Masciadri M, Pié J, Ramos FJ, Russo S, Selicorni A, Stefanova M, Strom TM, Werner R, Wierzba J, Zampino G, Gillessen-Kaesbach G, Wieczorek D, Kaiser FJ.
Expanding the clinical spectrum of the 'HDAC8-phenotype' - implications for molecular diagnostics, counseling and risk prediction.
Clin Genet. 2016 May;89(5):564-73. doi: 10.1111/cge.12717. Epub 2016 Jan 25.
2015
Parenti I, Gervasini C, Pozojevic J, Graul-Neumann L, Azzollini J, Braunholz D, Watrin E, Wendt KS, Cereda A, Cittaro D, Gillessen-Kaesbach G, Lazarevic D, Mariani M, Russo S, Werner R, Krawitz P, Larizza L, Selicorni A, Kaiser FJ.
Broadening of cohesinopathies: exome sequencing identifies mutations in ANKRD11 in two patients with Cornelia de Lange-overlapping phenotype.
Clin Genet. 2015 Feb 4. doi: 10.1111/cge.12564.
Braunholz D., Obieglo C., Parenti, I., Pozojevic J., Eckhold J., Reiz, B., Braenne I., Wendt KS, Watrin E., Vodopiutz J., Rieder H., Gillessen-Kaesbach G., Kaiser F.J.
Hidden Mutations in CdLS - Limitations of Sanger Sequencing in Molecular Diagnostics.
Hum Mutat. 2015 Feb;36(2):279-80.
2014
Kolovos P, van de Werken HJ, Kepper N, Zuin J, Brouwer RW, Kockx CE, Wendt KS, van IJcken WF, Grosveld F, Knoch TA.
Targeted Chromatin Capture (T2C): a novel high resolution high throughput method to detect genomic interactions and regulatory elements.
Epigenetics Chromatin. 2014 Jun 16;7:10. doi: 10.1186/1756-8935-7-10.
Wendt KS, Grosveld FG. Transcription in the context of the 3D nucleus. Curr Opin Genet Dev. 2014 Apr;25:62-7.
Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, Grosveld FG, Ren B, Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014 Jan 21;111(3):996-1001.
Braunholz D, Obieglo C, Parenti I, Pozojevic J, Eckhold J, Reiz B, Braenne I, Wendt KS, Watrin E, Vodopiutz J, Rieder H, Gillessen-Kaesbach G, Kaiser FJ. Hidden Mutations in CdLS - Limitations of Sanger Sequencing in Molecular Diagnostics. Hum Mutat. 2014 Sep 5. doi: 10.1002/humu.22685.
Kolovos P, van de Werken HJ, Kepper N, Zuin J, Brouwer RW, Kockx CE, Wendt KS, van IJcken WF, Grosveld F, Knoch TA. Targeted Chromatin Capture (T2C): a novel high resolution high throughput method to detect genomic interactions and regulatory elements. Epigenetics Chromatin. 2014 Jun 16;7:10.
2013
Zuin J, Franke V, van Ijcken WF, van der Sloot A, Krantz ID, van der Reijden MI, Nakato R, Lenhard B, Wendt KS. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 2014 Feb 13;10(2):e1004153. Epub Dec. 2013
Stadhouders R, Kolovos P, Brouwer R, Zuin J, van den Heuvel A, Kockx C, Palstra RJ, Wendt KS, Grosveld F, van Ijcken W, Soler E. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat Protoc. 2013 Mar;8(3):509-24.
2009
Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009 Nov;5(11):e1000739.
Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17(2):201-14.
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008 Feb 14;451(7180):796-801.
Bachelor and master projects
Cohesin functions and malfunctions
Genetic information in the form of the chromatin fiber is tightly packed into the nuclei of eukaryotic cells. To allow cellular functions the genetic information needs to be read out by the transcription machinery, duplicated by the replication machinery and the copies need to be identified and distributed correctly during cell division.
Cohesin is a ring-shaped protein complex critical to structure and handle the chromatin fiber during these processes. Defects in cohesin function can lead to chromosomal instability, gene expression defects and as consequence cancer and defects during embryonic development.
It is very important to understand the molecular mechanism of the cohesin complex to understand how mutations in the complex lead to disease. Great progress has been made in the past years and novel functions of cohesin have been uncovered. However, a number of fundamental questions are still unsolved. Some of these questions are outlined below.
If you look for an exciting project for your Bachelor and Master please contact us. The projects are suitable for students from the Molecular Medicine program, the Nanobiology program but also other master programs.
Bachelor and Master projects
Visualize cohesin dependent chromatin organization and respective changes after cohesin depletion using STORM and SIM imaging.
How many cohesin rings are necessary to form a loop?
How do the cohesin/CTCF binding sites that form the loop find each other?
According to the “loop extrusion model” cohesin actively pulls DNA through its protein ring. Can we prove/disprove this model?
During cell division chromatin is condensed into very compact metaphase chromosomes and cohesin is completely removed from chromatin. How are cohesin and CTCF sites reestablished after mitosis?
Cohesin & Disease – How do mutations observed in patients with severe developmental defects affect the functioning of the cohesin complex?
For details concerning the projects please contact k.wendt@erasmusmc.nl.
To approach these questions we use a wide panel of methodologies:
Imaging – Immunofluorescence staining, live cell imaging, STORM and SIM microscopy
Chromatin methods – ChIP, CHIP-sequencing and data analysis, chromatin conformation capturing techniques (4C, T2C, HiC)
Cell biology methods – CRISPR editing, manipulation of chromatin using dCAS9 (with tags), knockdown of proteins using siRNA
Biochemistry – Immunoprecipitation, Western blotting, antibody production and purification, mass spectrometry
Development of new methodology - Methods to study protein-chromatin interactions in a time-resolved manner
| Master projects: | Please inquire with us about current projects available |
| PhD positions: | None |
| Postdoc positions: | None |
Contact: Kerstin S. Wendt
Telephone: +31-107044007
E-mail: k.wendt@erasmusmc.nl
Group members
People:
Thomas van Staveren, PhD student
Macarena Moronta Gines, PhD student
Alternating master students and HBO students.

Kerstin S. Wendt
Telephone: +31-107044007
E-mail: k.wendt@erasmusmc.nl
Visiting address
Dept. of Cell Biology
Erasmus MC
Faculty building
Room Ee-1032
Wytemaweg 80
3015CN Rotterdam
The Netherlands
Mail address
Dept. of Cell Biology
Erasmus MC
Faculty building
Room Ee-1032
PO Box 2040
3000 CA Rotterdam
The Netherlands
