About Prof. dr. F.G. (Frank) Grosveld
Introduction
Regulation of transcription
The hematopoietic stem cell (HSC) is responsible for the formation of all blood cell lineages both in the embryo and the adult. HSC differentiation and lineage specification involve activation and repression of specific transcription programs. This is achieved by the coordinated action of conserved key transcription factors (TF). Our aim is to identify and characterize these TFs and to unravel their complex organization during blood cells differentiation. We also aim at understanding how erythroid genes get sequentially activated and repressed throughout development. Particular emphasis is given to the regulation of the β globin gene locus both as a model system to study the three dimensional organisation and interactions between regulatory regions at the molecular level and as a system that is responsible for β thalassemia and sickle cell anaemia. These two diseases are among the most widespread genetic diseases in the world and we are interested in exploiting the process of globin gene switching with the eventual aim to treat patients suffering from these diseases.
In order to achieve these goals we use a combination of state of the art technologies involving proteomic tools (protein tagging, single-step purification of TF complexes, mass spectrometry), genome-wide analysis of TF binding sites (chromatin immunoprecipitation coupled to high throughput sequencing, ChIP on chip, expression microarray analysis), RNAi library screens, and in vivo functional studies (mouse knock-in/knock out, morpholino-mediated knockdown of gene expression in zebrafish).
Contact information
Prof. Frank Grosveld
Department of Cell Biology
Dr. Molewaterplein 40
3015 GD Rotterdam
PO Box 2040
3000 CA Rotterdam
The Netherlands
e-mail : f.grosveld@erasmusmc.nl
Research projects
Hematopoietic transcription factor complexes
The hematopoietic stem cell (HSC) is responsible for the formation of all of the blood cell types. HSC differentiation involves coordinated and changing transcription, often by functionally conserved genes. For example, in mammals such a set of transcription factors (including Gata2, Tal1, Lmo2, Ldb1, Eto2, Gata1 and Runx1/Aml-1) is required for the differentiation of HSCs.
The laboratory has shown that each of these factors forms a number of different protein complexes that carry out different functions. For example the Gata1 TF forms a number of different complexes which can activate or repress transcription.
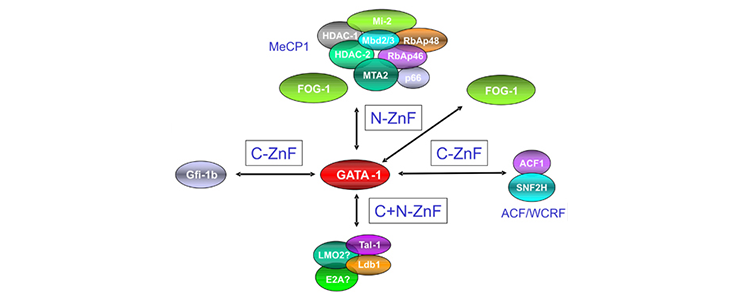
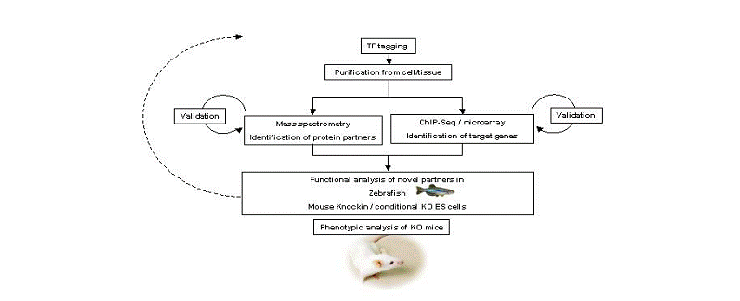
We performed single-step purification of transcription factor (TF) complexes in erythroid cells combined with mass spectrometry to characterize the composition of TF complexes. This approach led us to successfully identify new binding partners of essential hematopoietic TFs. These newly identified binding partners have essential roles in hematopoiesis as revealed by morpholino-mediated knock-down experiments in zebrafish embryo. We combine these data with genome-wide identification of TF binding sites using ChIP-Seq technology (chromatin immunoprecipitation coupled to high throughput sequencing) in order to map the different complexes to different gene loci. Taken altogether, these data lead us to build a comprehensive map of TF interactions and gene activation/repression during the course of hematopoietic cells differentiation.
Globin gene switching
The β globin genes are arranged in the same order in the β globin locus as their order of expression during development. The expression of the different genes at different stages is known as globin gene switching. The locus is characterized by a number of 3 dimensional interactions between different regulatory regions and these interactions change during development. β thalassemia and sickle cell anaemia which together are the most widespread genetic disease in the world, are usually characterized by mutations in or deletions of the adult β gobin gene, leaving the remainder of the locus unaltered. One of the primary aims of the laboratory is to understand the switch from the human foetal γ genes to the adult β gene with the aim to reverse this switch in patients. A number of different strategies are exploited to achieve this aim. In addition to the characterisation of transcription factor complexes (see above) a number of in vivo and functional approaches are also in progress. The first is crosslinking all of the protein complexes to the β globin promoter in vivo when the gene is silenced followed by the purification of the promoter and the characterisation of the factors by mass spectrometry. Subsequent functional studies will determine which of the factors are important for the silencing of the genes using mouse and zebrafish.
The second approach is purely based on function and involves a screen for β globin gene activation in reporter cells using shRNA libraries or heavy chain only antibody libraries. We are also collaborating with a number of laboratories to determine why patients react differently to hydroxyurea treatment using expression studies.
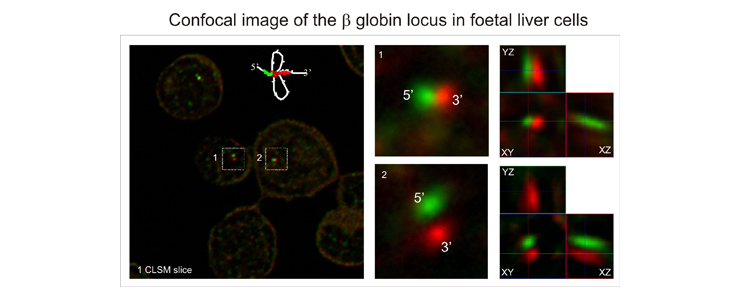
Finally we are interested in the three dimensional structure of complex gene loci in vivo and in particular that of the β globin locus as a model system. To that end we use 3C, 4C and ChIP methods in combination with confocal microscopy, 4pi microscopy and spatially modulated illumination (SMI) microscopy.
Publications
Significant publications from the last 5 years
Transcriptional Regulation by (Super)Enhancers: From Discovery to Mechanisms. Grosveld F., van Staalduinen J., Stadhouders R., Annu. Rev. Genom. Hum. Genet. (2021) in press.
A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Wang C, van Haperen R, Gutiérrez-Álvarez J, Li W, Okba NMA, Albulescu I, Widjaja I, van Dieren B, Fernandez-Delgado R, Sola I, Hurdiss DL, Daramola O, Grosveld F, van Kuppeveld FJM, Haagmans BL, Enjuanes L, Drabek D, Bosch BJ. Nat Commun. 2021 Mar 17;12(1):1715. doi: 10.1038/s41467-021-21968-w.PMID: 33731724
PLGA-Nanoparticles for Intracellular Delivery of the CRISPR-Complex to Elevate Fetal Globin Expression in Erythroid Cells. Cruz LJ, van Dijk T, Vepris O, Li TMWY, Schomann T, Baldazzi F, Kurita R, Nakamura Y, Grosveld F, Philipsen S, Eich C. Biomaterials. 2021 Jan;268:120580. doi: 10.1016/j.biomaterials.2020.120580.PMID: 33321292
An evolutionarily ancient mechanism for regulation of hemoglobin expression in vertebrate red cells. Miyata M, Gillemans N, Hockman D, Demmers JAA, Cheng JF, Hou J, Salminen M, Fisher CA, Taylor S, Gibbons RJ, Ganis JJ, Zon LI, Grosveld F, Mulugeta E, Sauka-Spengler T, Higgs DR, Philipsen S. Blood. 2020 Jul 16;136(3):269-278. doi: 10.1182/blood.2020004826.PMID: 32396940
A human monoclonal antibody blocking SARS-CoV-2 infection. Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus ADME, van Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch BJ. Nat Commun. 2020 May 4;11(1):2251. doi: 10.1038/s41467-020-16256-y.PMID: 32366817
Integrative and perturbation-based analysis of the transcriptional dynamics of TGFβ/BMP system components in transition from embryonic stem cells to neural progenitors. Dries R, Stryjewska A, Coddens K, Okawa S, Notelaers T, Birkhoff J, Dekker M, Verfaillie CM, Del Sol A, Mulugeta E, Conidi A, Grosveld FG, Huylebroeck D. Stem Cells. 2020 Feb;38(2):202-217. doi: 10.1002/stem.3111.
Investigation of the spatial structure and interactions of the genome at sub-kilobase-pair resolution using T2C. Kolovos P, Brouwer RWW, Kockx CEM, Lesnussa M, Kepper N, Zuin J, Imam AMA, van de Werken HJG, Wendt KS, Knoch TA, van IJcken WFJ, Grosveld F. Nat Protoc. 2018 Mar;13(3):459-477. doi: 10.1038/nprot.2017.132. Epub 2018 Feb 8.PMID: 29419817
PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cruz-Molina S, Respuela P, Tebartz C, Kolovos P, Nikolic M, Fueyo R, van Ijcken WFJ, Grosveld F, Frommolt P, Bazzi H, Rada-Iglesias A. Cell Stem Cell. 2017 May 4;20(5):689-705.e9. doi: 10.1016/j.stem.2017.02.004. Epub 2017 Mar 9.PMID: 28285903
Staff
Groupmembers
Rivan Cetin, PhD student
Ali Imam, senior scientist
Lucas Kuijpers, PhD student
Giulia Picco, PhD student
Jente van Staalduinen, PhD student
