What we do
About our project
Database
The shared database is password protected and accessible via the Internet for data entry, sharing and analysis. Online analysis tools are available:
- Incidence plots
- Pie charts
- Phylogenetic tree tool
- Geographical plot
he Noronet database can be accessed here.
Typing tool
In order to ensure standardized typing of the sequences, all submitted sequences are typed using the publicly accessible norovirus typing tool on the website of the RIVM.
Examples of what we do and newletters that you receive as a member
Below you can find an overview of the polymerase and capsid genotypes that members have reported to NoroNet for genogroup I (GI) and II (GII) from January 2015 until June 2018. Figures 1 to 7 show the data for the polymerase and capsid genotypes per year and per country. For GI we see co-circulation of several capsid and polymerase genotypes (Figure 1, Figure 5 and 7).
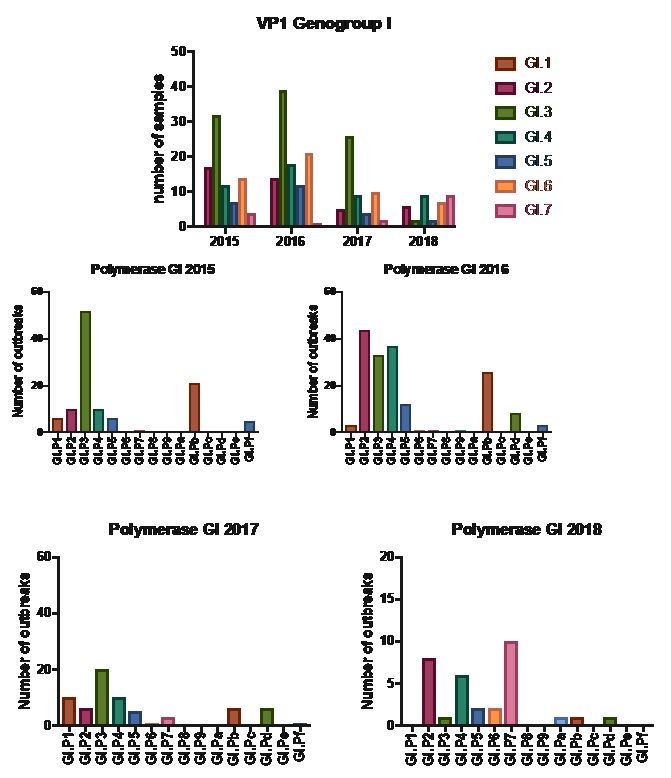
Figure 1 GI Polymerase and Capsid genotypes per year
For GII the most prevalent polymerase genotypes are GII.P4 Orleans 2009, GII.Pe, GII.P17 and GII.P16 (Figure 2, Figure 6). The polymerase GII.P16 has circulated for years with different capsid genotypes, but in the past years it became the predominant polymerase genotype and is mostly found in combination with the capsid genotypes GII.4 Sydney 2012 and GII.2 (Table 1.).
Although there are many GII capsid genotypes co-circulating there are large differences in prevalence. GII.4 variant GII.4 Sydney 2012 is still the most prevalent genotype (Figure 3, Figure 4 and Figure 7). For GII.17 there was a major increase in prevalence in 2015 and 2016, currently GII.17 outbreaks are still reported but at a lower frequency. For GII.2 there is also an increase in prevalence, mainly of GII.2 in combination with the GII.P16 genotype (Table 1).

Figure 2 Polymerase GII genotypes per year

Figure 3 GII Capsid genotypes per year
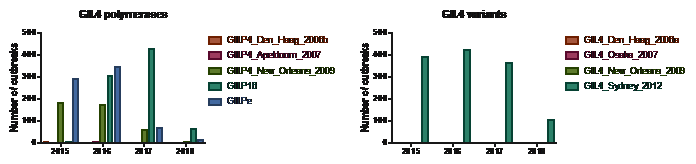
Figure 4 GII.4 variants, capsid and polymerases. *For GII.P16 all sequences were included including GII.P16 sequences that were detected in combination with a non-GII.4.
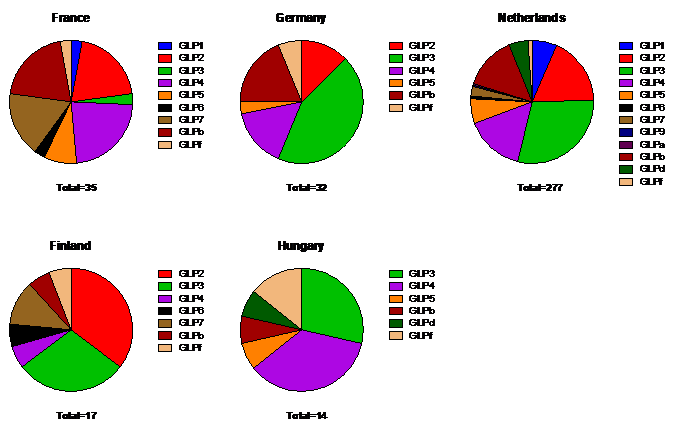
Figure 5. GI polymerase genotypes per country (>10 submitted sequences)
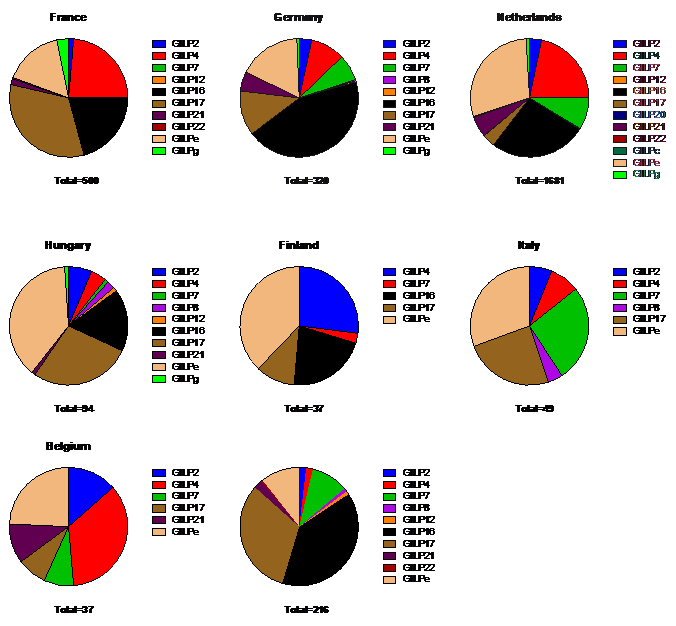
Figure 6. GII polymerase genotypes per country (>10 submitted sequences)
Our research focus
The aim of Noronet is to enlarge the knowledge on geographical and temporal trends in the emergence and spread of Norovirus variants, thus limiting the impact and scale of future norovirus epidemics. A second aim is the design of a well founded standardized nomenclature for existing and emerging norovirus genotypes and variants or sub-lineages. By prospectively sharing information on norovirus activity we can chart the global spread, recognize changes in circulating strains and possible changes in the epidemiology of the virus and thus potentially predict epidemic seasons.
Want to know more about the NoreoNet?
Contact Prof. Marion Koopmans: contact
Or visit www.noronet.nl
Collaborations
Members Global Noronet
Noronet has members in Australia, Austria, Belgium, Canada, Chile, China, Denmark, Finland, France, Germany, Hong Kong, Hungary, Ireland, Italy, Japan, Netherlands, New Zealand, Norway, Russia, Slovenia, South Africa, Spain, Sweden, United States and the United Kingdom
Publications
- Norovirus surveillance comes of age: the impact of NoroNet.
Green KY / Lancet Infect Dis. 2018 May;18(5):482-483. doi: 10.1016/S1473-3099(18)30062-8. Epub 2018 Jan 26. No abstract available / PMID:29396003 - Molecular surveillance of norovirus, 2005-16: an epidemiological analysis of data collected from the NoroNet network.
van Beek J, de Graaf M, Al-Hello H, Allen DJ, Ambert-Balay K, Botteldoorn N, Brytting M, Buesa J, Cabrerizo M, Chan M, Cloak F, Di Bartolo I, Guix S, Hewitt J, Iritani N, Jin M, Johne R, Lederer I, Mans J, Martella V, Maunula L, McAllister G, Niendorf S, Niesters HG, Podkolzin AT, Poljsak-Prijatelj M, Rasmussen LD, Reuter G, Tuite G, Kroneman A, Vennema H, Koopmans MPG; NoroNet.
Lancet Infect Dis. 2018 May;18(5):545-553. doi: 10.1016/S1473-3099(18)30059-8. Epub 2018 Jan 26 / PMID:29396001 - Global Spread of Norovirus GII.17 Kawasaki 308, 2014-2016
Chan MCW, Hu Y, Chen H, Podkolzin AT, Zaytseva EV, Komano J, Sakon N, Poovorawan Y, Vongpunsawad S, Thanusuwannasak T, Hewitt J, Croucher D, Collins N, Vinjé J, Pang XL, Lee BE, de Graaf M, van Beek J, Vennema H, Koopmans MPG, Niendorf S, Poljsak-Prijatelj M, Steyer A, White PA, Lun JH, Mans J, Hung TN, Kwok K, Cheung K, Lee N, Chan PKS. Emerg Infect Dis. 2017 Aug;23(8):1359-1354. doi: 10.3201/eid2308.161138 / PMID:28726618 - Emergence of a novel GII.17 norovirus – End of the GII.4 era?
de Graaf M, van Beek J, Vennema H, Podkolzin AT, Hewitt J, Bucardo F, Templeton K, Mans J, Nordgren J, Reuter G, Lynch M, Rasmussen LD, Iritani N, Chan MC, Martella V, Ambert-Balay K, Vinjé J, White PA, Koopmans MP. Euro Surveill. 2015 Jul 2;20(26). pii: 21178. PMID:26159308 - Norovirus genotype profiles associated with foodborne transmission, 1999-2012.
Verhoef L, Hewitt J, Barclay L, Ahmed SM, Lake R, Hall AJ, Lopman B, Kroneman A, Vennema H, Vinjé J, Koopmans M. Emerg Infect Dis. 2015 Apr;21(4):592-9. doi: 10.3201/eid2104.141073. PMID:25811368 - Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012.
van Beek J, Ambert-Balay K, Botteldoorn N, Eden JS, Fonager J, Hewitt J, Iritani N, Kroneman A, Vennema H, Vinjé J, White PA, Koopmans M; NoroNet. Euro Surveill. 2013 Jan 3;18(1):8-9. PMID:23305715 - Proposal for a unified norovirus nomenclature and genotyping.
Kroneman A, Vega E, Vennema H, Vinjé J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. Arch Virol. 2013 Oct;158(10):2059-68. doi: 10.1007/s00705-013-1708-5. Epub 2013 Apr 25. PMID:23615870 - An automated genotyping tool for enteroviruses and noroviruses.
Kroneman A, Vennema H, Deforche K, v d Avoort H, Peñaranda S, Oberste MS, Vinjé J, Koopmans M. J Clin Virol. 2011 Jun;51(2):121-5. doi: 10.1016/j.jcv.2011.03.006. Epub 2011 Apr 21. PMID:21514213



